stylised-example
cynthiahqy
2022-11-10
Last updated: 2022-11-22
Checks: 7 0
Knit directory: viz-panel-maps/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221110) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cb702ba. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: README_files/
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: analysis/figure/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/stylised-example.Rmd) and
HTML (docs/stylised-example.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | cb702ba | cynthiahqy | 2022-11-22 | add sketch example, fix renv dependencies |
| html | cb702ba | cynthiahqy | 2022-11-22 | add sketch example, fix renv dependencies |
| html | 1b832de | cynthiahqy | 2022-11-10 | Build site. |
| Rmd | 68db7fa | cynthiahqy | 2022-11-10 | add stylised example starting with matrix cases |
| html | 68db7fa | cynthiahqy | 2022-11-10 | add stylised example starting with matrix cases |
knitr::opts_chunk$set(message = FALSE)Define and visualise concordances
Consider the following (weighted) concordance between lower case and upper case letters (representing categories) represented in the following forms:
as Matrix
library(Matrix)
## define source (x-to-x) cases
one_to_one <- diag(1, 2, 2)
many_to_one <- matrix(
c(1, 1, 1, 1, 0, 0,
0, 0, 0, 0, 1, 1), ncol = 2)
one_to_many <- matrix(
c(0.5, 0.5, 0, 0, 0, 0, 0, 0.3, 0.3, 0.4), nrow = 2,
byrow=TRUE
)
## combine into incidence matrix
b_mtx <- Matrix::bdiag(many_to_one, one_to_one, one_to_many)
dimnames(b_mtx) <- list(letters[1:nrow(b_mtx)],
paste0(LETTERS[1:ncol(b_mtx)], LETTERS[1:ncol(b_mtx)])
)
print(b_mtx)10 x 9 sparse Matrix of class "dgCMatrix"
AA BB CC DD EE FF GG HH II
a 1 . . . . . . . .
b 1 . . . . . . . .
c 1 . . . . . . . .
d 1 . . . . . . . .
e . 1 . . . . . . .
f . 1 . . . . . . .
g . . 1 . . . . . .
h . . . 1 . . . . .
i . . . . 0.5 0.5 . . .
j . . . . . . 0.3 0.3 0.4as iGraph Object
Forms a bipartite graph as target and source classification are disjoint sets.
library(igraph, quietly = TRUE)
## convert matrix to graph
b_igraph <- igraph::graph_from_incidence_matrix(b_mtx,
directed = TRUE,
mode = "out",
weighted = TRUE)
print(b_igraph)IGRAPH 3961ec4 DNWB 19 13 --
+ attr: type (v/l), name (v/c), weight (e/n)
+ edges from 3961ec4 (vertex names):
[1] a->AA b->AA c->AA d->AA e->BB f->BB g->CC h->DD i->EE i->FF j->GG j->HH
[13] j->IIplot(b_igraph, layout = layout_as_bipartite)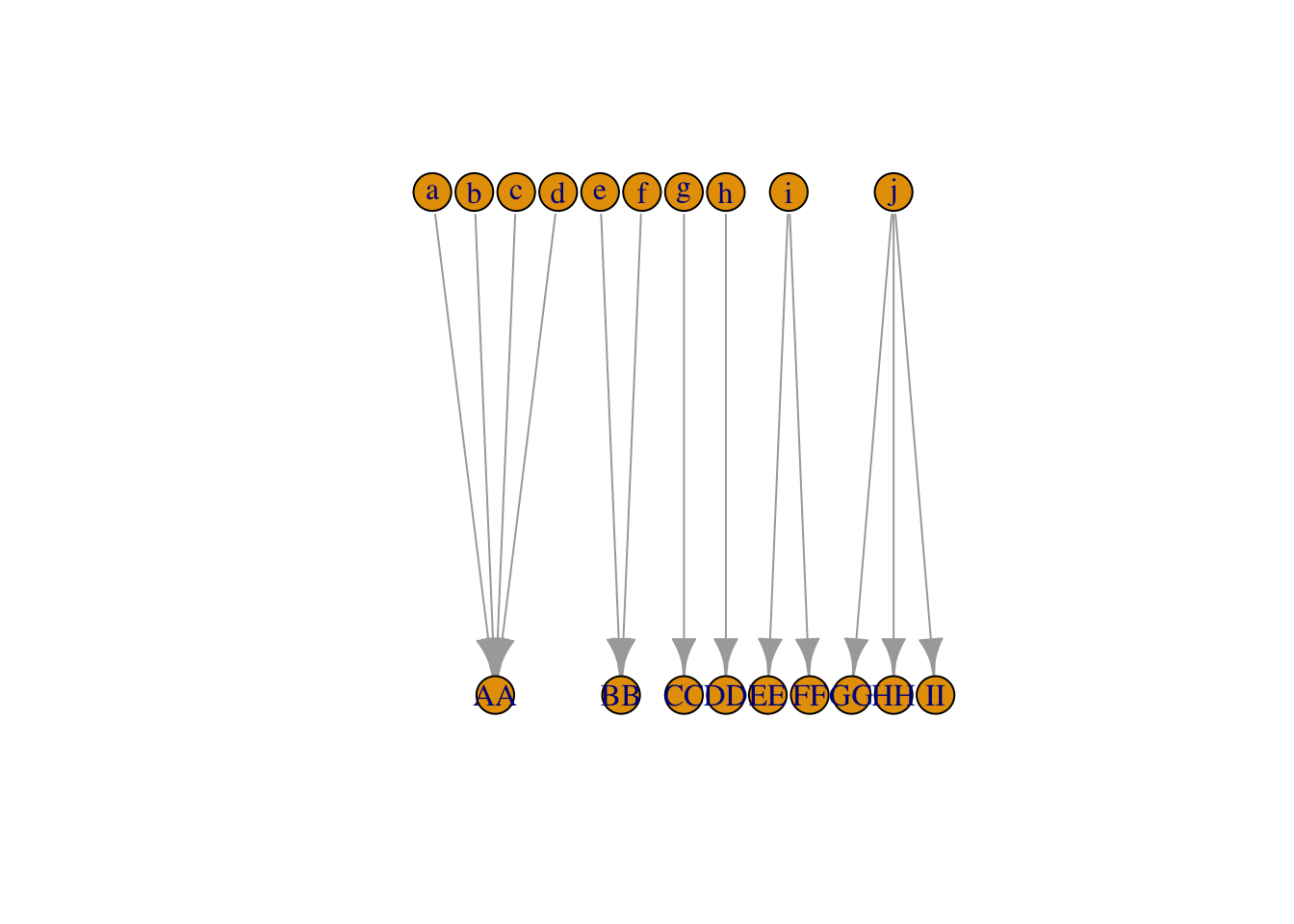
| Version | Author | Date |
|---|---|---|
| 68db7fa | cynthiahqy | 2022-11-10 |
as tidygraph Object
library(ggraph, quietly = TRUE)
library(tidygraph, quietly = TRUE)
## convert from igraph
b_tgraph <- tidygraph::as_tbl_graph(b_igraph)
print(b_tgraph)# A tbl_graph: 19 nodes and 13 edges
#
# A rooted forest with 6 trees
#
# Node Data: 19 × 2 (active)
type name
<lgl> <chr>
1 FALSE a
2 FALSE b
3 FALSE c
4 FALSE d
5 FALSE e
6 FALSE f
# … with 13 more rows
#
# Edge Data: 13 × 3
from to weight
<int> <int> <dbl>
1 1 11 1
2 2 11 1
3 3 11 1
# … with 10 more rows## plot using ggraph
ggraph(b_tgraph, layout = "bipartite") +
geom_edge_diagonal() +
geom_node_label(aes(label=name)) +
scale_y_reverse() +
scale_x_reverse() +
coord_flip()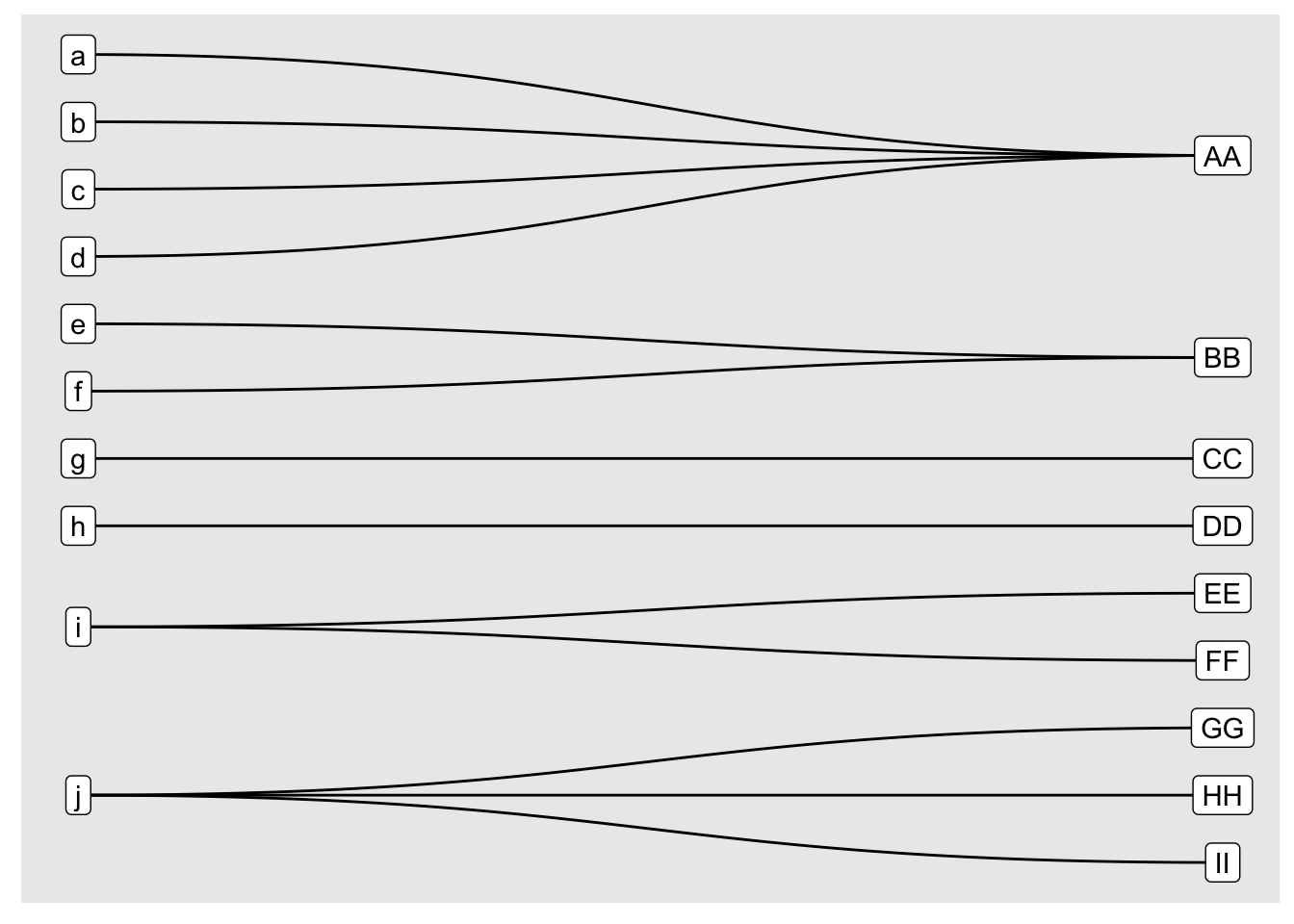
| Version | Author | Date |
|---|---|---|
| 68db7fa | cynthiahqy | 2022-11-10 |
Weighted concordance as Panel Map
conformr’s panel_map class expects the
weighted concordance to be provided in this form,
## using iGraph
b_pm <- igraph::get.data.frame(b_igraph)
print(b_pm) from to weight
1 a AA 1.0
2 b AA 1.0
3 c AA 1.0
4 d AA 1.0
5 e BB 1.0
6 f BB 1.0
7 g CC 1.0
8 h DD 1.0
9 i EE 0.5
10 i FF 0.5
11 j GG 0.3
12 j HH 0.3
13 j II 0.4Notice the first two columns of the dataframe form the edge list of the graph, and the third column stores the weights of each edge
## plot using ggplot and ggbump
source(here::here('R/plt_pm_df.R'))
plt_pm_df(b_pm, from, to, weight)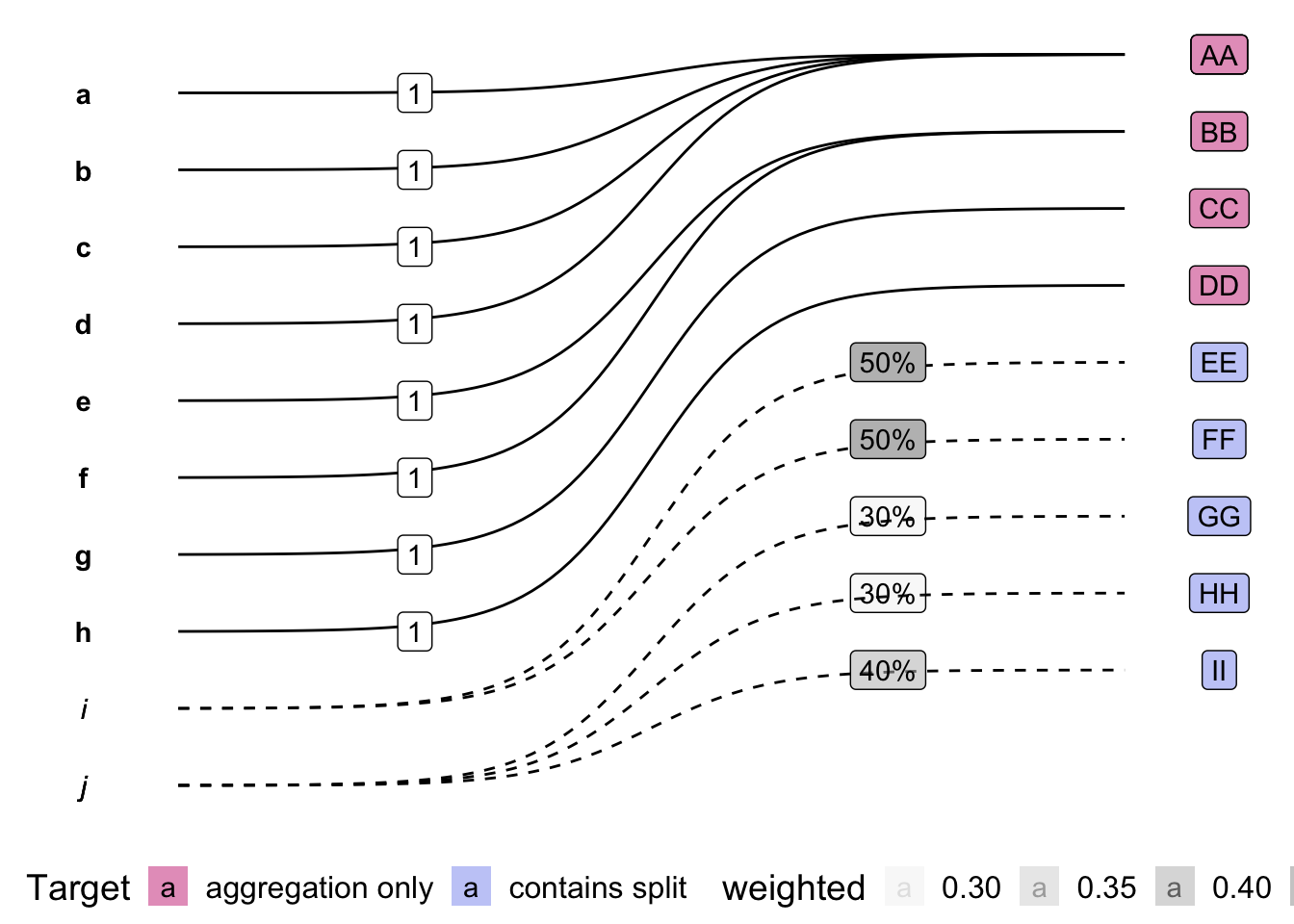
| Version | Author | Date |
|---|---|---|
| 68db7fa | cynthiahqy | 2022-11-10 |
Using Panel Maps to Transform Data
in Matrix Notation
- Let \(\bf{B}\) be a \(n \times m\) matrix, satisfying the necessary conditions to form a panel map
- Let \(\bf{x}\) be vector of length \(n\) containing the numeric values to be transformed.
- Then the transformed data is \(\bf{y} = \bf{B}'\bf{x}\)
x <- rep_len(100, nrow(b_mtx))
y <- t(b_mtx) %*% x
y9 x 1 Matrix of class "dgeMatrix"
[,1]
AA 400
BB 200
CC 100
DD 100
EE 50
FF 50
GG 30
HH 30
II 40using {conformr}
data_in <- tibble::tibble(from = rownames(b_mtx), gdp_in = x)
# conformr::validate_panel_map(b_pm) |>
# conformr::validate_coverage(b_pm, data, values_from, from_code, to_code, weights) |>
conformr::use_panel_map(map = b_pm, data=data_in, values_from=gdp_in,
from_code = from, to_code = to,
weights = weight, .suffix = "_out")# A tibble: 9 × 2
to gdp_in_out
<chr> <dbl>
1 AA 400
2 BB 200
3 CC 100
4 DD 100
5 EE 50
6 FF 50
7 GG 30
8 HH 30
9 II 40Conditions for a valid transformations
A: valid-weights
The sum of weights associated with a given source node/class must sum to one. If weights total less than one, some portion of the initial source value will be “lost” in the transformation. Conversely, if the weights exceed one, then some part of the initial source value is being “double” transferred.
Violations of this condition:
as Matrix Row totals
- Let \(\bf{Z}\) be a \(n \times m\) matrix showing the incidence
between two disjoint sets of nodes (e.g
from/to) - Then \(\bf{Z}\) is a valid panel map if and only if \(\bf{Z}\boldsymbol{1} = \boldsymbol{1}\) where \(\boldsymbol{1}\) is a unit vector of length \(m\)
## define source (x-to-x) violations
one_to_more <- c(0.5, 0.7)
one_to_less <- c(0.3, 0.4)
wgt_mtx <- bdiag(one_to_one, t(one_to_more), t(one_to_less))
source(here::here("code/fnc-make-dimnames.R"))
dimnames(wgt_mtx) <- make_dimnames(wgt_mtx)
wgt_mtx4 x 6 sparse Matrix of class "dgCMatrix"
AA BB CC DD EE FF
a 1 . . . . .
b . 1 . . . .
c . . 0.5 0.7 . .
d . . . . 0.3 0.4## validation check
(sum_wgt_mtx <- wgt_mtx %*% rep_len(1, ncol(wgt_mtx)))4 x 1 Matrix of class "dgeMatrix"
[,1]
a 1.0
b 1.0
c 1.2
d 0.7## problem source nodes
wgt_mtx[!as.vector(sum_wgt_mtx == 1), ]2 x 6 sparse Matrix of class "dgCMatrix"
AA BB CC DD EE FF
c . . 0.5 0.7 . .
d . . . . 0.3 0.4as Data Frame Group totals
The data frame equivalent of the matrix rows total calculation involves grouping the non-trival edges (i.e. weights are non-zero), and summarising the total weights for each group. Any groups with total weights not equal to one require correction.
## convert violation matrix back to data frame
mtx <- wgt_mtx
wgt_df <- mtx |>
as.data.frame.array() |>
tibble::as_tibble(rownames = "from") |>
tidyr::pivot_longer(cols=c(-from), values_to="weights", names_to="to") |>
dplyr::filter(weights!=0)
## calculate total weights for each source node
wgt_df |>
dplyr::group_by(from) |>
dplyr::summarise(t_weights = sum(weights))# A tibble: 4 × 2
from t_weights
<chr> <dbl>
1 a 1
2 b 1
3 c 1.2
4 d 0.7A*: valid-edgelist
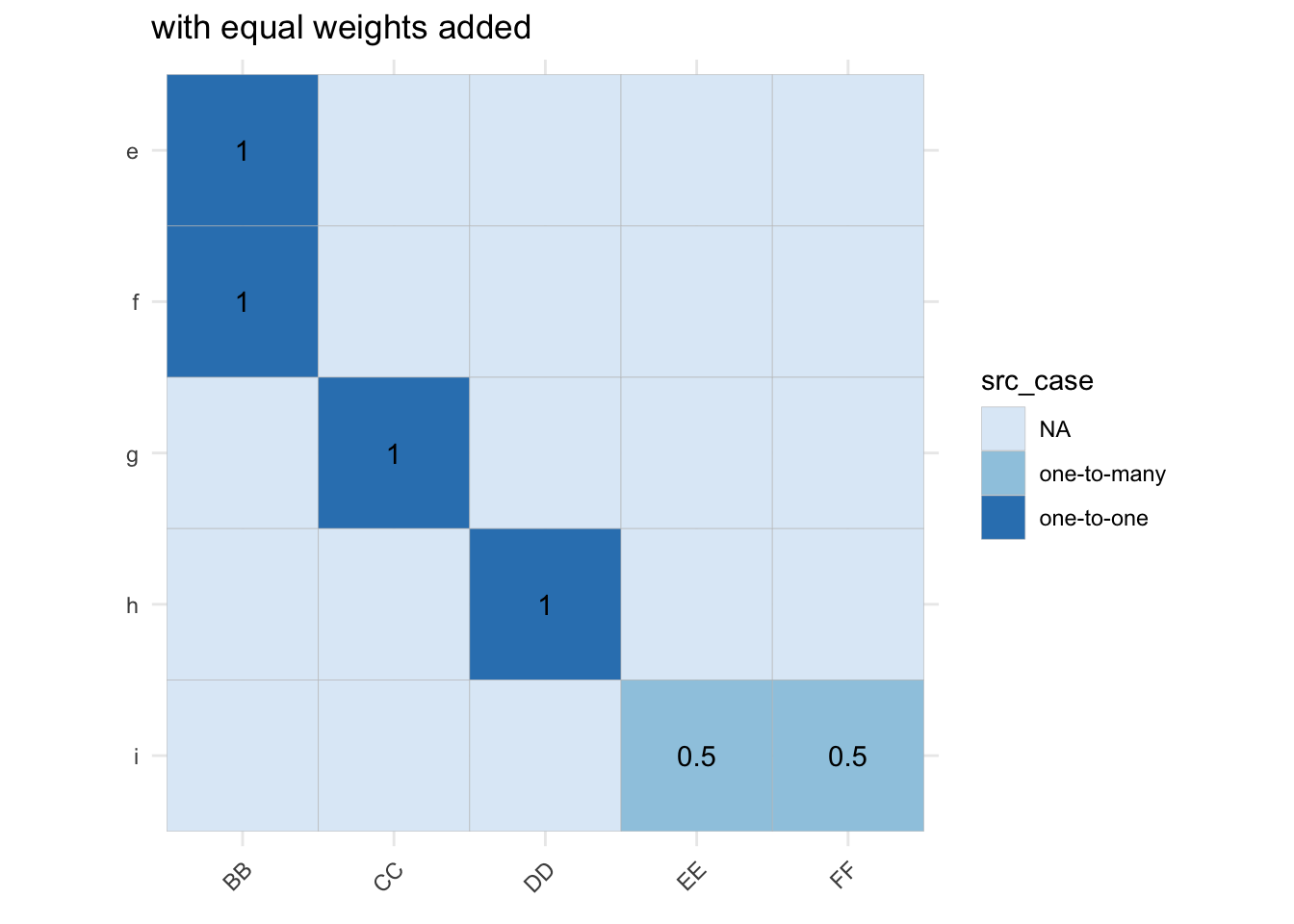
The make_panel_map_equal()1 function is basically
a helper which assigns equal fractional weights to links between source
and target nodes.
b_mtx_long <- b_mtx[-c(1:4, 10), -c(1, 7:9)] |>
as.data.frame.array() |>
tibble::as_tibble(rownames = "from") |>
tidyr::pivot_longer(cols=c(-from), values_to="weight", names_to="to")
gg <- b_mtx_long |>
dplyr::mutate(src_case = dplyr::case_when(weight==1 ~ "one-to-one",
weight==0 ~ "NA",
weight < 1 ~ "one-to-many")) |>
ggplot(aes(x=to, y=from)) +
geom_tile(aes(fill=src_case), col="grey") +
scale_y_discrete(limits=rev) +
scale_fill_brewer() +
coord_fixed() +
labs(x = element_blank(), y = element_blank()) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
ggtitle("concordance table as matrix")
gg_weights <- gg + geom_text(data = dplyr::filter(b_mtx_long, weight !=0), aes(label=weight)) +
ggtitle("with equal weights added")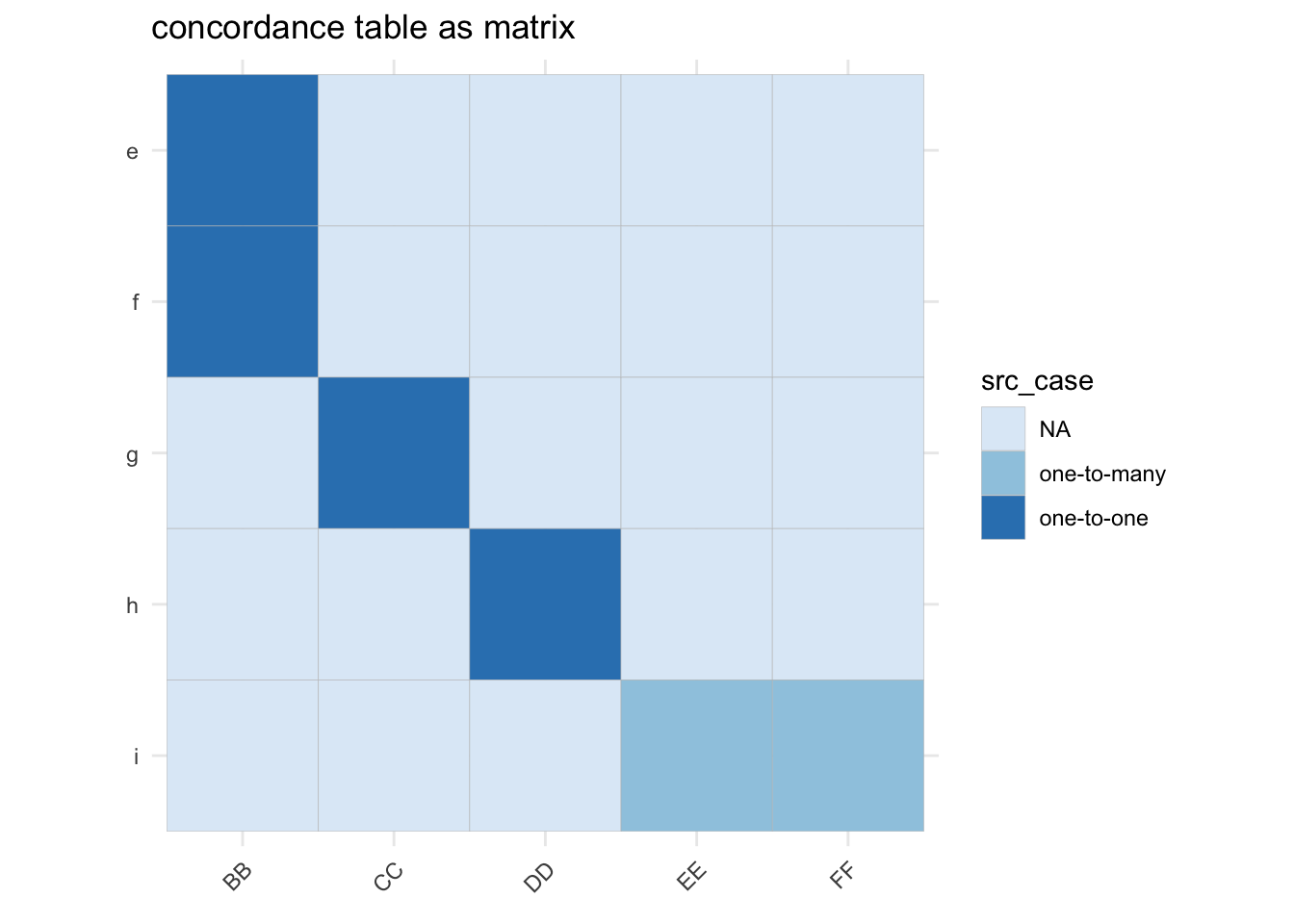
| Version | Author | Date |
|---|---|---|
| cb702ba | cynthiahqy | 2022-11-22 |
| Version | Author | Date |
|---|---|---|
| cb702ba | cynthiahqy | 2022-11-22 |
A necessary condition for this helper to work as intended is that the list of un-weighted edges doesn’t have any duplicates.
B: complete-coverage
Conformability is a necessary but not sufficient condition for a valid transformation.
The necessary condition is that the Panel Map used in the transformation covers all the Source Codes present in the Source Data. When applying the transformation using table joins, the extraneous codes are dropped and conformability is implicitly achieved.
as Matrix conformability condition
## add an extra source class
bad_x <- rep_len(100, nrow(b_mtx) + 1)
## now we cannot use the panel map
t(b_mtx) %*% as.matrix(bad_x)Error in t(b_mtx) %*% as.matrix(bad_x): Cholmod error 'X and/or Y have wrong dimensions' at file ../MatrixOps/cholmod_sdmult.c, line 88as Set comparison
C: complete-data-in
NA values should be dealt with explicitly before applying a panel map transformation.
Properties of valid transformations
If all three conditions (A, B, C) are met, then:
- sum of
value_inandvalue_outwill be equal - OTHER SMELL CHECKS??
ASIDE: Converting between Representations
PM <–> Matrix <– iGraph –> PM
panel map to Matrix
source(here::here('R/pm_to_matrix.R'))
(pm2mtx <- pm_to_matrix(b_pm, to=to, weights=weight))10 x 9 sparse Matrix of class "dgCMatrix"
AA BB CC DD EE FF GG HH II
a 1 . . . . . . . .
b 1 . . . . . . . .
c 1 . . . . . . . .
d 1 . . . . . . . .
e . 1 . . . . . . .
f . 1 . . . . . . .
g . . 1 . . . . . .
h . . . 1 . . . . .
i . . . . 0.5 0.5 . . .
j . . . . . . 0.3 0.3 0.4panel map to iGraph (broken)
Vertex metadata errors can be avoided by going from data frame to matrix first (as seen above)
## reconstruct node/vertices list
b_pm_nodes <- tidyr::pivot_longer(b_pm[, 1:2], from:to, names_to = "type", values_to = "name") %>%
dplyr::distinct() %>%
dplyr::arrange(type) %>%
as.data.frame()
rownames(b_pm_nodes) <- b_pm_nodes$name
b_pm2igraph <- igraph::graph_from_data_frame(b_pm, directed = TRUE, vertices = b_pm_nodes)Error in igraph::graph_from_data_frame(b_pm, directed = TRUE, vertices = b_pm_nodes): Duplicate vertex namesplot(b_pm2igraph, layout = layout_as_bipartite)Error in plot(b_pm2igraph, layout = layout_as_bipartite): object 'b_pm2igraph' not foundigraph to long data frame
b_igraph |> igraph::as_long_data_frame() from to weight from_type from_name to_type to_name
1 1 11 1.0 FALSE a TRUE AA
2 2 11 1.0 FALSE b TRUE AA
3 3 11 1.0 FALSE c TRUE AA
4 4 11 1.0 FALSE d TRUE AA
5 5 12 1.0 FALSE e TRUE BB
6 6 12 1.0 FALSE f TRUE BB
7 7 13 1.0 FALSE g TRUE CC
8 8 14 1.0 FALSE h TRUE DD
9 9 15 0.5 FALSE i TRUE EE
10 9 16 0.5 FALSE i TRUE FF
11 10 17 0.3 FALSE j TRUE GG
12 10 18 0.3 FALSE j TRUE HH
13 10 19 0.4 FALSE j TRUE II
sessionInfo()R version 4.2.1 (2022-06-23)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur ... 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] ggbump_0.1.0 dplyr_1.0.10 tidygraph_1.2.2 ggraph_2.1.0
[5] ggplot2_3.3.6 igraph_1.3.5 Matrix_1.4-1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ggrepel_0.9.1 Rcpp_1.0.9 here_1.0.1
[4] lattice_0.20-45 tidyr_1.2.1 getPass_0.2-2
[7] ps_1.7.1 assertthat_0.2.1 rprojroot_2.0.3
[10] digest_0.6.30 utf8_1.2.2 ggforce_0.4.1
[13] wesanderson_0.3.6 R6_2.5.1 evaluate_0.16
[16] httr_1.4.3 highr_0.9 pillar_1.8.1
[19] rlang_1.0.6 data.table_1.14.2 rstudioapi_0.13
[22] whisker_0.4 callr_3.7.0 jquerylib_0.1.4
[25] rmarkdown_2.14 labeling_0.4.2 stringr_1.4.1
[28] polyclip_1.10-4 munsell_0.5.0 compiler_4.2.1
[31] httpuv_1.6.5 xfun_0.31 pkgconfig_2.0.3
[34] htmltools_0.5.2 tidyselect_1.2.0 tibble_3.1.8
[37] gridExtra_2.3 graphlayouts_0.8.3 viridisLite_0.4.1
[40] fansi_1.0.3 withr_2.5.0 later_1.3.0
[43] MASS_7.3-57 grid_4.2.1 jsonlite_1.8.2
[46] gtable_0.3.1 lifecycle_1.0.3 DBI_1.1.3
[49] git2r_0.30.1 magrittr_2.0.3 scales_1.2.1
[52] cli_3.4.1 stringi_1.7.8 conformr_0.0.0.9001
[55] farver_2.1.1 renv_0.15.5 viridis_0.6.2
[58] fs_1.5.2 promises_1.2.0.1 bslib_0.3.1
[61] ellipsis_0.3.2 generics_0.1.3 vctrs_0.4.2
[64] cowplot_1.1.1 RColorBrewer_1.1-3 tools_4.2.1
[67] glue_1.6.2 tweenr_2.0.2 purrr_0.3.5
[70] processx_3.6.1 fastmap_1.1.0 yaml_2.3.5
[73] colorspace_2.0-3 knitr_1.39 sass_0.4.2.9000 other possible names:
equal_pm_from_nodes()↩︎